research
Main menu
Molecular automata
Molecular automata assembly: principles and simulation of bacterial membrane construction
In order to understand the basic rules and principles governing molecular self-
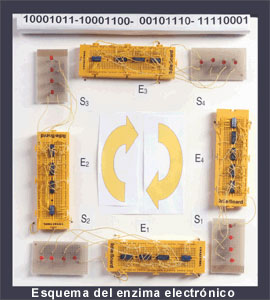
'Evolving hardware' as model of enzyme evolution
Organism growth
and survival is based on thousands of enzymes organized in networks.
The motivation to understand how a large number of enzymes evolved so
fast inside cells may be relevant to explaining the origin and
maintenance of life on Earth. We introduced
electronic circuits called ‘electronic enzymes’ that model
the catalytic function performed by biological enzymes [14].
Electronic enzymes are the hardware
realization of enzymes defined as molecular automata with a finite
number of internal conformational states and a set of Boolean operators
modelling the active groups of the active site. One of the main
features of electronic enzymes is the possibility of evolution finding
the proper active site by means of a genetic algorithm yielding a
metabolic ring or k-
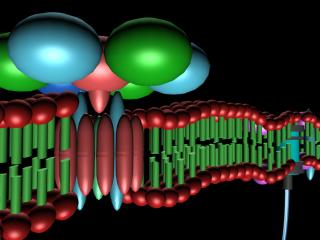
Molecular automata modeling in structural biology
Dynamic
activities within living cells rest on biomolecular systems organized
into cellular structures and organelles. A common motivation of
computer simulation in the past decade has been to understand cellular
complexity by developing models from which to derive powerful unifying
generalizations and predictions of cell dynamics. However, the modeling
and simulation of cell dynamics present a host of theoretical and
practical challenges. These challenges involve the need to achieve some
level of competence in cellular and molecular principles (i.e.,
enzymology, polymerization, self-